How to find putative genes of interest
One of the outputs of the Tomato Expression Atlas is the Expression Cube which displays a list of genes grouped by their similarity in expression profiles. Co-expressed genes tend to participate in the same biological processes and therefore this co-expression tool can be used to reveal new genes potentially involved in similar processes as the query gene. As explained in previous sections, pasting the query gene ID into the input box named By Gene ID and clicking on Get Expression will display the expression values from the query gene and a list of candidates genes with correlated expression profiles.
4.1 Example: Identification of candidate genes involved in vascular tissue development
As an example, we will use the gene Solyc04g080940, with the description "Noduline-like protein", as a query gene. This gene is homolog of the Medicago truncatula gene "nodulin 21", which is upregulated in Rhizobium-induced Medicago nodules (Ranocha et al. 2013), and to the Arabidopsis thaliana gene AT1G75500, "Walls are thin 1" (WAT1), a member of the plant drug/metabolite exporter (P-DME) family. WAT1 is preferentially expressed in vascular tissues, including developing xylem vessels and fibers (Ranocha et al. 2010), and WAT1 mutants show defects in cell elongation, a dwarfed phenotype and little to no secondary cell walls in stem fibers (Ranocha et al. 2010). WAT1 has also been shown to act as a vacuolar auxin transport facilitator required for auxin homoeostasis and involved in regulating fibre cell wall thickness (Ranocha et al. 2013).
The tomato WAT homolog, Solyc04g080940 (Figure 1) is highly expressed in the pericarp vascular tissue during cell expansion,
from 5 days post anthesis (DPA) to the mature green stage.
The list of genes co-expressed with Solyc04g080940 suggests genes with functions related to WAT1 in vascular tissue development and hormone/amino acids transport.
Particularly interesting are Solyc01g100920 and Solyc03g005480, which are also described as "Noduline-like proteins" (table 1, in yellow):
and homologs to arabidopsis genes encoding amino acids transporters (TAIR). Other genes involved on vascular tissue development (xylem/phloem)
that are shown in the co-expression cube are the genes Solyc03g111810, Solyc05g013850 and Solyc05g013870 (table 1, in gray).
These genes are homolog of AT3G01680, which encodes a structural protein found in phloem sieve elements (Anstead et al. 2012, Ruping et al. 2010).
The list of co-expressed genes in the expression cube can indicate candidate genes potentially involved in the same biological process or pathway,
including transcription factors that may regulate their expression, and that can then be selected for experimental validation.
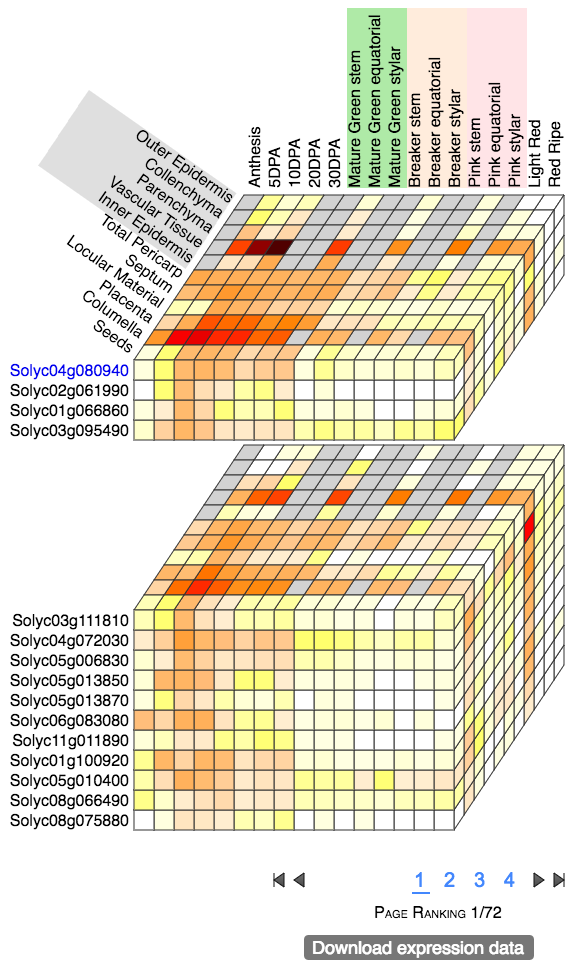
Figure 1. Expression Cube output for Solyc04g080940.
| Gene ID | Description | Correlation val. | Ath Homolog | Ath Gene Name |
|---|---|---|---|---|
| Solyc04g080940 | Nodulin-like protein | 1 | AT1G75500 | WAT1/UMAMIT5 |
| Solyc02g061990 | Basic leucine zipper transcription factor | 0.91 | AT2G17770 | ATBZIP27 |
| Solyc01g066860 | Unknown Protein | 0.9 | AT1G63310 | |
| Solyc03g095490 | Receptor like kinase, RLK | 0.9 | AT5G58300 | |
| Solyc03g111810 | Sieve element-occluding protein 3 | 0.9 | AT3G01680 | AtSEOR1 |
| Solyc04g072030 | Interferon-induced GTP-binding protein Mx | 0.9 | AT1G60500 | DRP4C |
| Solyc05g006830 | Thioredoxin H | 0.9 | AT5G39950 | ATTRX2 |
| Solyc05g013850 | Sieve element-occluding protein 3 | 0.9 | AT3G01680 | AtSEOR1 |
| Solyc05g013870 | Sieve element-occluding protein 3 | 0.9 | AT3G01680 | AtSEOR1 |
| Solyc06g083080 | Unknown Protein | 0.9 | AT5G48500 | |
| Solyc11g011890 | Zinc finger protein 7 | 0.9 | AT1G66140 | ZFP4 |
| Solyc01g100920 | Nodulin-like protein | 0.89 | AT1G09380 | UMAMIT25 |
| Solyc05g010400 | Receptor like kinase, RLK | 0.89 | AT5G16000 | ATNIK1 |
| Solyc08g066490 | Receptor like kinase, RLK | 0.89 | AT2G25790 | SKM1 |
| Solyc08g075880 | Metal ion binding protein | 0.89 | AT2G18196 | |
| Solyc09g009830 | Carbonic anhydrase (Carbonate dehydratase) | 0.89 | ||
| Solyc11g011920 | Glutamate decarboxylase | 0.89 | ||
| Solyc11g069570 | Cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG | 0.89 | ||
| Solyc01g101120 | Glucan endo-1 3-beta-glucosidase 1 | 0.88 | ||
| Solyc02g076690 | Cathepsin B-like cysteine proteinase | 0.88 | ||
| Solyc02g080900 | Genomic DNA chromosome 5 TAC clone K6A12 | 0.88 | ||
| Solyc03g005480 | Nodulin-like protein | 0.88 | AT2G39510 | UMAMIT14 |
References
| Anstead et al. 2012 | Arabidopsis P-Protein Filament Formation Requires Both AtSEOR1 and AtSEOR2 |
| Ranocha et al. 2010 | Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers |
| Ranocha et al. 2013 | Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis |
| Ruping et al. 2010 | Molecular and phylogenetic characterization of the sieve element occlusion gene family in Fabaceae and non-Fabaceae plants |




